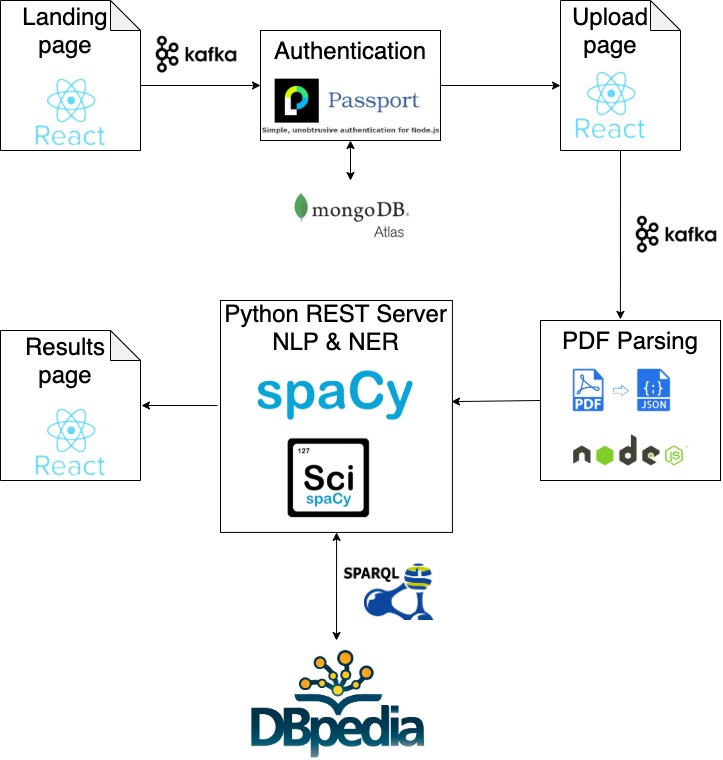
How I created a scalable Kafka powered React web app that uses NLP to understand medical reports!
Description
The application called MediReport enables end-users, who do not have a medical background, to interpret the meaning of various medical/clinical terms that are present in their medical reports. By querying more than 9.5 billion RDF triples[1] in DBpedia, I was able to access a huge structured dataset of limitless information.
The application provides the users with personalized results about their medical conditions, which can be translated into multiple languages.
Approach
- Provide PDF upload feature for medical reports via React-based UI/Node.js
- Use pdf2json to extract text data from the PDF medical report
- Use spaCy and scispaCy to perform NER to extract medical terms
- Enhance the above-extracted data by providing explanations of each term using DBpedia
- Use Transformers NLG library to generate a human-like response for the user.
Application flow
Results page
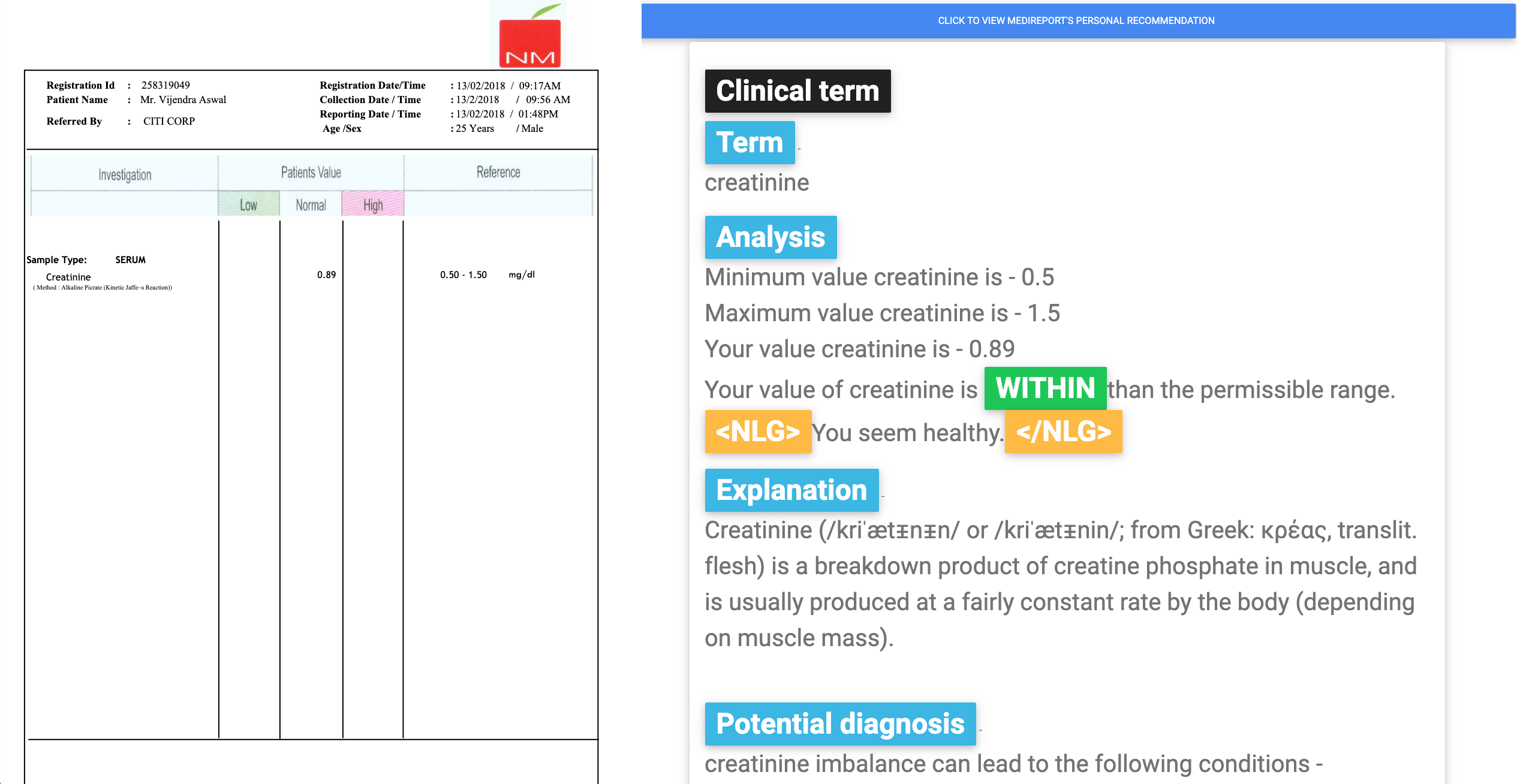
It contains two React components - the image of the uploaded PDF and the results after parsing the PDF, performing NER and getting data from DBpedia.
<div className='rowC'>
<div style=>
<Document
file={this.state.file}
onLoadSuccess={this.onDocumentLoadSuccess}
>
<Page pageNumber={this.state.pageNumber}/>
</Document>
Page {this.state.pageNumber} of {this.state.numPages}
</div>
<div style=>
<Button
onClick={() => this.setState({isOpenModal: true})}
type="button" variant="primary">
Click to view MediReport's personal recommendation
</Button>
{this.populateSection()}
</div>
</div>
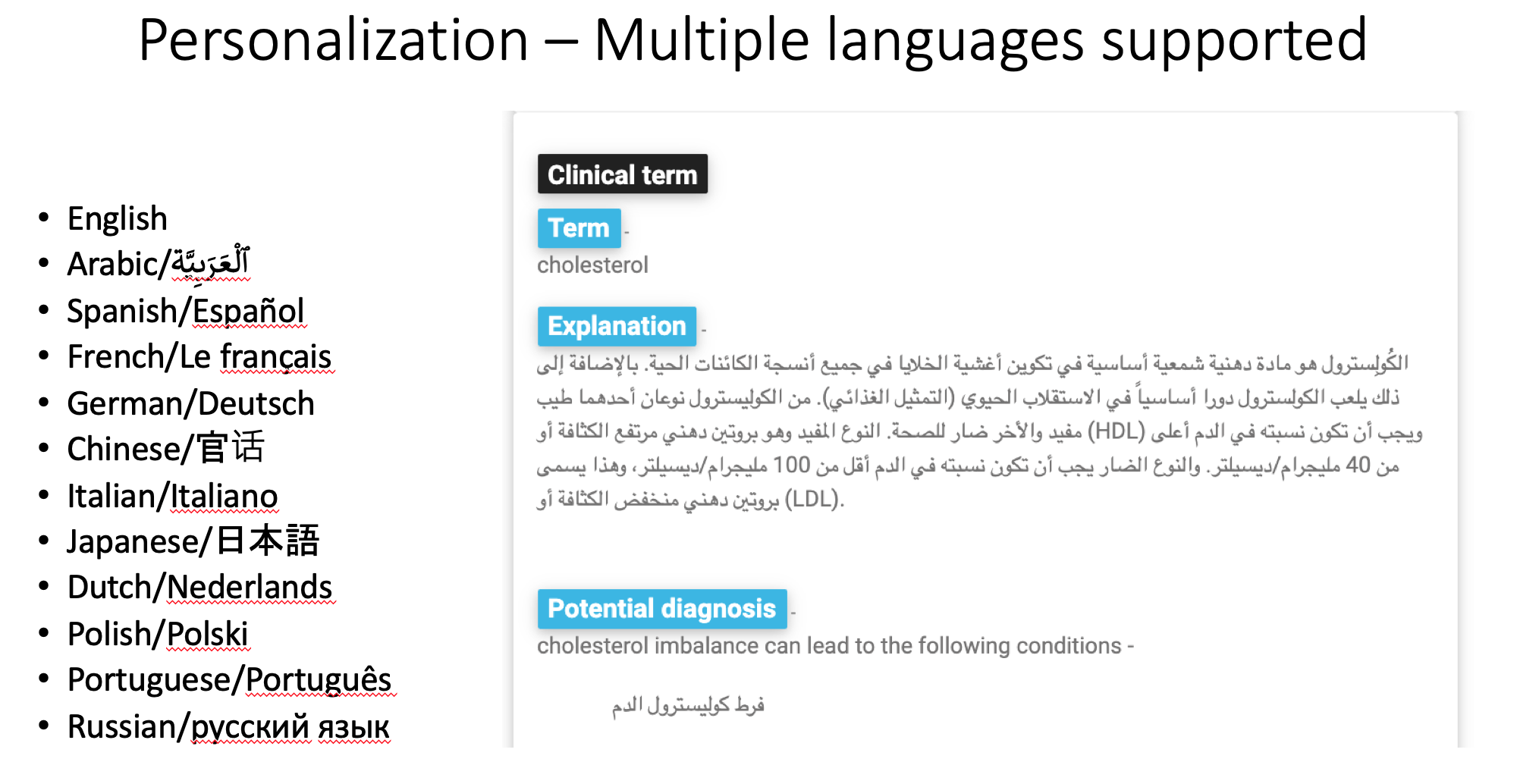
Because I was querying a huge multi-lingual database, I could easily change the language of the text at the click of one the language buttons.

Using Kafka to scale
Apache Kafka® is a distributed streaming platform that is used to publish and subscribe to streams of records, similar to a message queue or enterprise messaging system. Since all messages and REST calls are stored in Kafka topics, the application can scale without having to worry about durability and fault-tolerance.
For example, to scale my login functionality, I created a Kafka topic named ‘access’ using the below command -
bin/kafka-topics.sh --create --zookeeper localhost:2181 --replication-factor 2 --partitions 2 --topic access
Hence all login requests, instead of being served immediately, are stored in the topics ‘access’ as can be seen below. So even when I do not have the compute capacity to process millions of requests immediately or my application crashes, I will not lose any request since Kafka replicates them to multiple partitions and keeps track of the messages that have already been processed using a technique called commit offset.
router.post('/login', auth.optional, function (req, res) {
kafka.make_request('access', {"path": "signin", "body": req.body}, function (err, result) {
console.log('in result');
console.log(result);
if (err) {
res.send({
signinSuccess: false,
signinMessage: "Sign In Failed"
})
} else {
res.send(result);
}
});
});
All other REST calls (sign-up, upload PDF, process PDF, etc.) were similarly stored in Kafka other topics.
The Python NLP server and clinical NER
While Flask is one of the more popular web application frameworks for Python, I used Klein to create my Python REST server because of its ease of use.
class ItemStore(object)
app = Klein()
def _init_(self):
self._items = {}
@app.route('/<string:name>', methods=['POST'])
def post_item(self, request, name):
request.setHeader('Content-Type', 'application/json')
content = json.loads(request.content.read())
Once I had my POST REST call to accept parsed PDF data, I performed the following NLP operations -
-
Performed pre-processing to remove whitespaces, linebreaks, all non-alphanumeric characters and commonly occurring redundant data such as “gm”, “mg/dl”, etc. which were leading to low NER recall value.
text = text\ .replace("\r", " ")\ .replace("\n", " ") \ .replace("gm", " ") \ .replace("mg/dl", " ")\ .lower() text = re.sub(r'[^A-Za-z ]+', " ", text) text = re.sub(' +', " ", text) -
Performed lemmatization on every word in the text to group all the inflected forms so that they can be analyzed as a single item. For example, ‘pain’, ‘pains’ and ‘paining’ will be mapped to the lemma - ‘pain’.
sp = spacy.load('en_core_web_sm') sentence = sp(text) text = " ".join([token.lemma_ for token in sentence]) -
Performed NER using pre-trained scispaCy model ‘en_ner_bc5cdr_md’
nlp = spacy.load("en_ner_bc5cdr_md") doc = nlp(json.dumps(text)) entities = doc.ents
Querying billions of DBpedia triples
Each entity extracted in the previous step is considered to be a potential answer and hence is supplied as a parameter.
def getQueryResults(self, entitySet, langCode):
data = {'entities': []}
for entity in entitySet:
result_entity = {"entityName": "", "comment": "", "foods": [], "diseases": []}
abstract_sparql_query = self.getAbstractQuery(entity, langCode)
abstract_query_results = abstract_sparql_query.query().convert()
For example, when ‘cholesterol’ is passed into the query function in the variable ‘entity’, it is used to generate a SPARQL query which runs on DBpedia’s cloud endpoint - http://dbpedia.org/sparql.
def getAbstractQuery(self, entity, langCode):
sparql = SPARQLWrapper("http://dbpedia.org/sparql")
sparql.setReturnFormat(JSON)
query = """
PREFIX dbo: <http://dbpedia.org/ontology/>
PREFIX dbp: <http://dbpedia.org/resource/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT distinct(?chem) ?comment where
{
?chem rdf:type dbo:ChemicalCompound .
?chem rdfs:label ?label .
?chem rdfs:comment ?comment .
FILTER regex(?label, "^%s$", "i")
FILTER (langMatches(lang(?comment),"%s"))
}
""" % (entity, langCode)
sparql.setQuery(query)
return sparql
langCode is ‘en’ or English by default but can be one of the following values based on what the user selects.
const enNameToLanguageCodeMap = new Map([
["English", "en"],
["Arabic", "ar"],
["Spanish", "es"],
["French", "fr"],
["German", "de"],
["Chinese", "zh"],
["Italian", "it"],
["Japanese", "ja"],
["Dutch", "nl"],
["Polish", "pl"],
["Portuguese", "pt"],
["Russian", "ru"],
]);
Technologies used
Programming Languages: Javascript, HTML5, SPARQL, Python
Web/Mobile Frameworks: React, Node.js, Redux, Passport, Bootstarp
Databases: MongoDB, DBpedia
Cloud Technologies: Kafka, AWS EC2, Docker, Kafka
References
- YEAH! We did it again ;) – New 2016-04 DBpedia release. DBpedia. 19 October 2016.